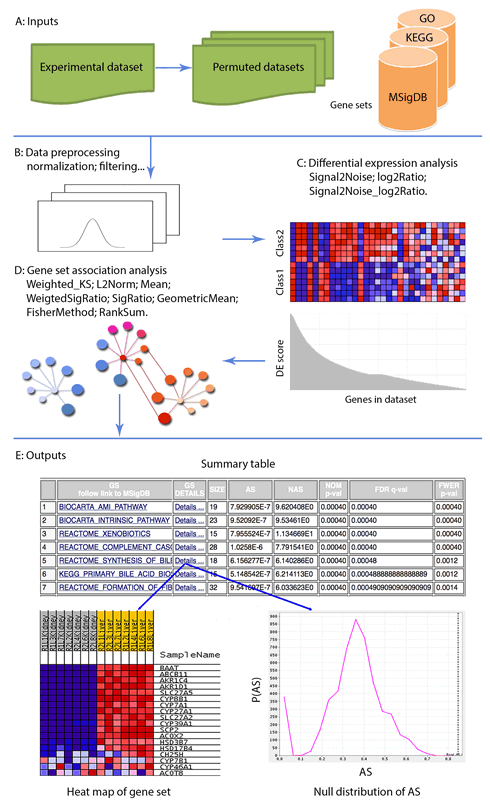
overview
Gene Set Association Analysis for RNA-Seq (GSAASeqSP and GSAASeqGP) are toolsets for gene set association analysis of RNA-Seq count data. GSAASeqSP and GSAASeqGP identify pathways/gene sets significantly associated with a disease or a phenotype by analyzing genome-wide patterns of gene expression variation measured by RNA-Seq technology. Both sample/phenotype permutation and gene permutation are implemented in GSAASeqSP while GSAASeqGP is purely based on gene permutation. We recommend using GSAASeqSP with permutation type set to "phenotype" (default) when there are at least 7 samples in each phenotype, otherwise using GSAASeqSP with permutation type set to "gene_set" or GSAASeqGP.
Gene Set Association Analysis (GSAA) is a Bioinformatics platform for integrative gene set association analysis of SNP data and microarray gene expression data. GSAA identifies pathways/gene sets significantly associated with a disease or a phenotype through integrating evidence from genome-wide patterns of genetic variation and gene expression variation of two phenotypes.
Gene Set Association Analysis-SNP (GSAA-SNP) is a Bioinformatics platform for gene set association analysis of SNP data. GSAA-SNP identifies pathways/gene sets significantly associated with a disease or a phenotype by analyzing genome-wide patterns of genetic variation of two phenotypes.
what's new
October 2, 2015: GSAA 2.0 has just been released. In this new version, the non-directional association analysis has been changed to directional analysis for all methods in GSAASeqSP. A gene set may be "up-regulated" or "down-regulated" relative to a particular phenotypic class.
September 12, 2015: GSAA 1.5 has just been released. 2 new human gene set collections (Wikipathways and Pathway Interaction Database collections) and 12 Arabidopsis gene set collections have been added to the gene set database in this version. Human gene sets were collected by Dr. William Ackerman at The Ohio State University Wexner Medical Center. Arabidopsis gene sets were extracted from the AraPath database (http://bioinformatics.sdstate.edu/arapath/).
July 4, 2015: GSAA 1.3 has just been released. This new version fixed a problem on the FTP connection and is much faster in loading the genesets database.
August 10, 2014: Simulated RNA-Seq count datasets are now available. These datasets were designed for evaluating and comparing the performance of set/pathway based approaches.
October 20, 2013: A new version of gene set association analysis (GSAA 1.2) has just been released. This new release includes gene set association analysis for RNA-Seq with sample permutation (GSAASeqSP).
May 20, 2012: The version 1.1 of gene set association analysis (GSAA 1.1) is now available. This new release includes gene set association analysis for RNA-Seq with gene permutation (GSAASeqGP).
September 22, 2011: The first public release of gene set association analysis (GSAA) is now available. GSAA is developed for joint gene set association analysis based on both microarray gene expression data and SNP data.
September 22, 2011: The first public release of gene set association analysis-SNP (GSAA-SNP) is now available. GSAA-SNP is developed for gene set association analysis solely based on SNP data.
September 22, 2011: Simulated microarray gene expression datasets, SNP datasets, and gene set datasets are now available. These datasets were designed for evaluating the performance of set/pathway based approaches.
Getting Started
Documentation: The documentation includes tutorials and user guides that provide general information and instructions with regard to preparing data, running analyses, and interpreting results for GSAA, GSAA-SNP, GSAASeqSP, and GSAASeqGP.
Software: The implementations of GSAA, GSAA-SNP, GSAASeqSP, and GSAASeqGP.
Datasets: Simulated RNA-Seq count data, microarray gene expression data, SNP data, and gene set data.
Registration
The GSAA, GSAA-SNP, GSAASeqSP, and GSAASeqGP are free for non-commercial use. If you are an academic user, click here to register, and then proceed to download. Registration is free and easy.